In this post I will look at cython for speeding up python code. This is my first attempt at using cython so please bear with me.
Installation
Cython itself can be installed straight forward with pip:
pip install cython
In order to compile the python code to c you will need a c compiler. Depending on your platform you will already have one (e.g. gcc on linux). As I am on a windows environment I can just mingw or one of the visual studio compilers. I chose to use mingw in this case.
You can download mingw from this link: https://sourceforge.net/projects/mingw/files/. When installing it is enough to choose the mingw32-base option.
After installing we need to add the mingw/bin folder to the path.
We also need to add distutils.cfg file in our python environment base path. In my case that is located at:
C:\Users\\AppData\Local\Programs\Python\Python36
As content to the file we will add:
[build]
compiler = mingw32
[build_ext]
compiler = mingw32
We are now ready to start using cython!
Let's have a look at performance
As an example I will look at two of the implementations of the fibonacci sequences from the post Calculating the last digits of large fibonacci numbers. Calculating the last digits of large fibonacci numbers
It is worth noting that the fibonacci numbers are perhaps not the best example, both in terms of code, but also as one would have to take extreme care when dealing with larger numbers where integer overflows and the like becomes a real issue.
Let's first take the implementations of the fibonacci sequence and store them in fibonacci_py.py.
def fib1(n):
if n <= 1:
return n
else:
return fib1(n - 1) + fib1(n - 2)
def fib3(n):
if n == 0:
return 0
f1 = 0
f2 = 1
for a in range(n - 1):
f1, f2 = f2, (f1 + f2)
return f2
Let's set up a small test script that we will reuse later to time the various implementations:
import timeit
fib1 = min(timeit.repeat('fibonacci_py.fib1(30)', setup='import fibonacci_py', repeat=10, number=1))
fib3 = min(timeit.repeat('fibonacci_py.fib3(30)', setup='import fibonacci_py', repeat=10, number=1))
base = fib1
print("Function \t Speedup")
print("fib1 (naïve) \t {0:f}".format(base / fib1))
print("fib3 (linear) \t {0:f}".format(base / fib3))
This outputs:
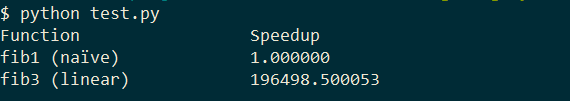
Let's just say, the linear version is faster.
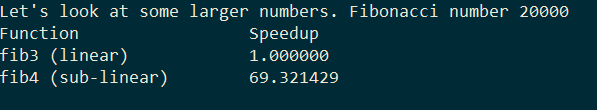
Ona side note, the sub-linear version mentioned in the previous post is much faster for large fibonacci numbers:
Making use of cython<
On of the great things about cython is that you can start of with regular python code and iteratively add more and more type information and write more c-like code as you go.
Let's start by taking the code exactly like it is of the fib1 function and saving it in a file called fibonacci_cy.pyx.
In order to build the c-code we will create a setup.py file and add the following:
from distutils.core import setup
from Cython.Build import cythonize
setup(ext_modules=(cythonize("fibonacci_cy.pyx")))
Now we can build it by executing python setup.py build_ext --inline. You will see a lot of output from the compiler and it will leave you with a file called something along the lines of fibonacci_cy.cp36-win_amd64.pyd (.so on linux). This file can be imported as normal in python using import fibonacci_cy.
Let's setup a small test script again to test the performance. Let's call it test_cy_vs_py.py:
import timeit
fib1 = timeit.timeit('fibonacci_py.fib1(30)', setup='import fibonacci_py', number=20)
fib1_cy = timeit.timeit('fibonacci_cy.fib1(30)', setup='import fibonacci_cy', number=20)
base = fib1
print("Function\t Speedup")
print("fib1 (naïve) \t {0:f}".format(base / fib1))
print("fib1_cy (naïve) \t {0:f}".format(base / fib1_cy))
This outputs:
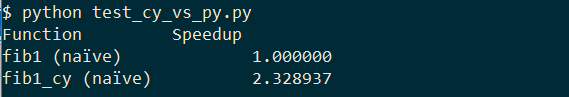
We have more than 2x speed up without changing any code at all. Let's help the cython compiler out by providing some type information in fibonacci_cy.py:
cpdef int fib1(n):
if n <= 1:
return n
else:
return fib1(n - 1) + fib1(n - 2)
We run the test again and get:
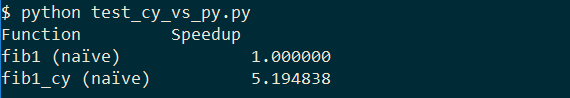
So just by specifying the return type we get a speedup of 5x. Not much more to do here, but we can also specify which type n has:
cpdef int fib1(int n):
if n <= 1:
return n
else:
return fib1(n - 1) + fib1(n - 2)
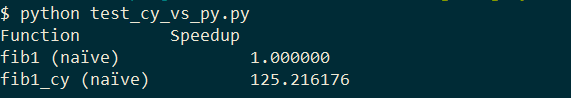
So by adding just the return and input type we achieve a ~100x speed up. Not bad :)
Doing the same exercise for the fib3 function we achieve ~20x speed up, by just defining type information:
cpdef int fib3(int n):
if n == 0:
return 0
cpdef int f1 = 0
cpdef int f2 = 1
for a in range(n - 1):
f1, f2 = f2, (f1 + f2)
return f2
output:
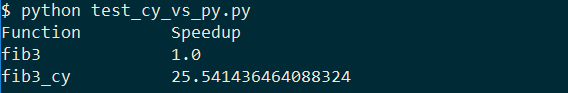
That is it for now. I will post more on cython as I get a bit more experience with the tool and use it in real life applications.
Tip: you can pass annotate=True to the cythonize function in setup.py. This will produce an html file showing which lines are still using python code and which are converted to c-code. This is a great way to find which lines can still be optimized. More on this in a coming blog post.